DNA is the very reason you exist the way you do. It is the backed up files from your family tree with the information on how to make the proteins that make you. Today, DNA can be read, made, downloaded and installed. This concept is what created much of the biotech industry as we know it today. But how do you do this? (not the figurative ‘you’, actual YOU) I bet you didn’t know that just about anyone can go online, design and order DNA, as well as all the chemical reagents and tools needed to put that DNA in whatever living thing you can think up. There are some really clever techniques and methods of doing so that people like you are employing to chase their biological dreams.
What are BioBricks?
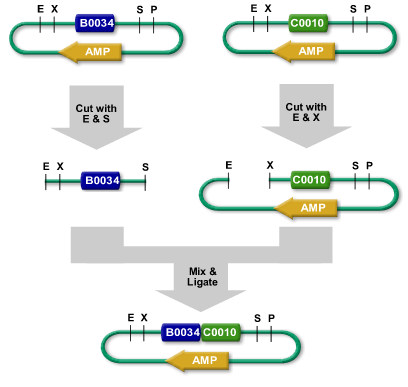
BioBricks are exactly what they sound like, they are essentially the LEGOs of genetic information. They are small pieces of DNA isolated and categorized by their function. They are cut using a standardized set of restriction enzymes which target certain nucleotide sequences and cut at those spots, what are called restriction sites. After they cut, they leave an open ended sequence, also called a “sticky” end, which, like LEGO, can be easily assembled with other pieces of DNA.
Restriction enzymes were discovered in bacteria as a means of defending themselves from bacteriophages, which are like viruses for bacteria. The first discovered and a commonly used restriction enzyme is EcoR1 (Eco = E.coli, R1 = first restriction enzyme).
What are Plasmids, Operons, and Regulatory Sequences?
The information these BioBricks encode are broken down into a few main categories. First, to understand these sequences, we have to understand how genes are coded and decoded in the cell. DNA is read by three bases at a time, these are called codons. All encoding sequences are made of codons, but how does the transcriptase know where to start and stop reading the DNA?
An example of the Biobrick symbols and construct design. Source
There are specific codons for starting and stopping that the enzyme can detect. These are just the basics, there are also sequences that certain proteins called inhibitors bind to. Where they bind is larger than a single codon and is meant to block transcription unless acted upon by the right ligand/activity. Along with inhibitors, there are promoters that attract transcription agents to specific areas. These too have activation/repression binding capabilities. Operons are another name for these clustered regulatory sequences, consisting of a promoter, a terminator and an operator. When exploring biobricks you can browse all these various genetic switches and triggers to add to your construct.
Constructs and Plasmid Design
When I say construct, I mean specially a designed genetic code, consisting of only a few protein coding sequences, whether it’s a pathway or just the genes for expressing one protein. Typically, this refers to plasmids but it doesn’t always have to. Since some genetic constructs are used in their linear form or integrated into genomes. Building standardized collections of constructs is a way to test parts against each other. For instance, if you have GFP (green fluorescent protein) coding sequences in plasmids with different operons or promoters, you will see a difference in expression.
In mammals and plants, these collections of DNA sequences are how our bodies determine what a tissue makes/does. Because the constructs in our genome are turned on in some tissues and not in others, you can take a cheek cell and a piece of hair and find the exact same DNA (your entire genome) in both. Understanding these constructs, and how we can build them is also important in understanding how stem cells work. All cells were once stem cells and just specified through necessity and biological interactions, and not necessarily by definitive design.
How to Get Started
Now BioBricks aren’t free to anyone. BioBricks are made for the iGEM competition, which isn’t hard to enter, and I believe there is a lab fee involved. Mind you, the cost is nothing compared to what it would cost to sequence and synthesize all of those components yourself. There are also plenty of videos online on how to splice together your own BioBrick plasmids. Now, depending on your project, BioBricks might not be your best bet considering they do have limited parts and are mostly designed for yeast and E. coli, at the moment. One important thing you need is some software to help you out. There are plenty of programs that will be willing to sort through your genes and help you design a construct that you can then order from a manufacturer.
BioBrick Alternatives
Don’t have a sequenced gene of interest? No access to BioBricks or no BioBricks to suit your purpose? Know of the organism the genes you want come from? My suggestion is to use cDNA. cDNA is DNA made from RNA. With this technique, it is easy to isolate protein coding sequences by sampling mRNA and using a reverse transcriptase to turn it into cDNA that you can then separate out in a gel, and ligate to plasmid backbone with selectable markers. With the right precautions, steps, and knowhow, you can isolate your own bricks to use in your constructs. You just need a means of identifying what gene your plasmid is expressing. This is ideal for obviously expressed genes like pigments, smells, or resistances (like antibiotic resistance). Just make sure you understand the pathway and what you want from a tissue, and that you understand whatever organism you test the cDNA in.
Other at Home Methods
In my next posts, I will be showcasing more methods of genetic experimentation that may be more easy to follow, fun and affordable. Also on December 9th my animated DIY Biohacking video will be released make sure to keep a lookout for this exciting new content. If you have any comments or questions, leave them in the comments below I would love to be able to respond.